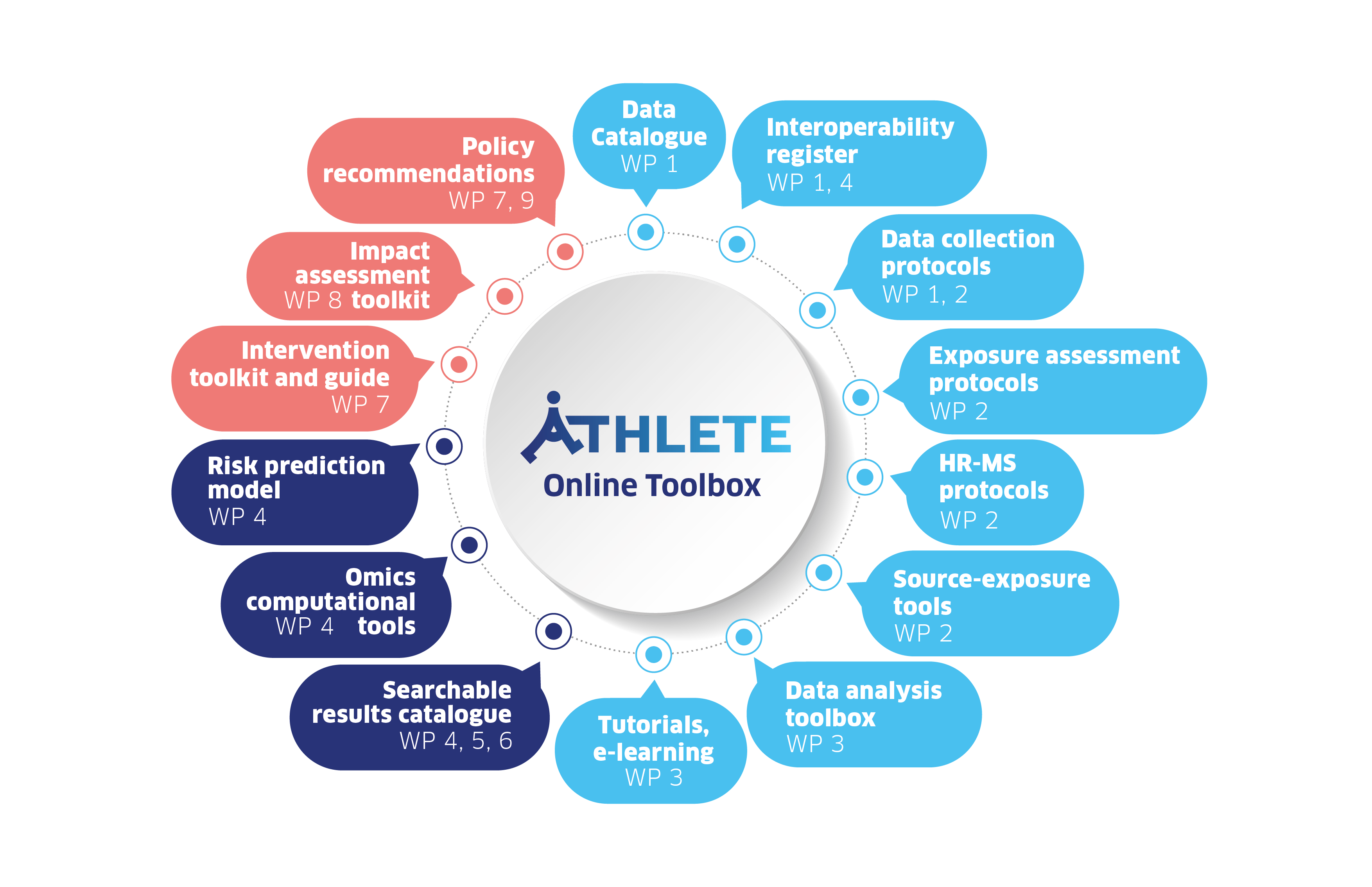
Toolbox
All parts of the work will provide input to the ATHLETE Online Toolbox on this page. This will ensure that data and tools will not only be developed and used within the project, but will be accessible to researchers and policy makers long after the project has finished.
The figure on the right shows the contributions of each Work Packages (WP) to the online toolbox.
HELIX Project
HELIX Project
The HELIX project represents a collaborative study across six established and ongoing longitudinal population-based birth cohort studies in six European countries (France, Greece, Lithuania, Norway, Spain, and the United Kingdom).
Exposome data
European Networks Health Data and Cohort Catalogue
This catalogue contains metadata on cohorts/data sources, the variables they collect, and/or harmonization efforts to enable integrated reuse of their rich valuable data.
Data access protocols
This tool is a protocol, or technical explanation / ‘manual’ for implementation and maintenance of the federated data access system which has been put in place by WP1. The intended audience are members of technical teams within institutions taking -or wanting to take- part in the Athlete research network.
Protocol and SOPs for exposure data, health outcome data and biological samples collection in adolescents (HELIX Subcohort)
This tool present the protocol and SOPs for the collection of data on cardiometabolic, respiratory and neurodevelopment/mental health outcomes, exposures data, and biological samples collection for the follow-up of the HELIX Subcohort cohort into adolescence.
Exposure Assessment
Exposure assessment protocols
This document describes the protocols for ‘Personal monitoring data in cohort and intervention study subjects’ (Task 2.3.2) and ‘Assessing the external exposome’ (Task 2.4).
EXPOApp3 and EXPOHub for personal monitoring of physical activity and localization
The smartphone app ExpoApp3 and its software platform (EXPOHub) have been developed by the SME Bettair Cities within ATHLETE. The app monitors, in real-time, location (GPS), accelerometer, physical activity and screen (on/off). The app can be used alone or as part of a larger set of data collection tools (e.g. wearable and air pollution monitoring devices) to record personal exposure data in epidemiological and intervention studies.
Analysis Tools
Exposome data analysis tutorial
A colaboratory project combining both “plain text” explanations and “R code chunks” for running analyses. Explanations include different exposome analysis steps, namely 1) descriptive analysis, 2) Visualization, and 3) Association analysis. This material was generated in Spanish and targeted researchers in environmental epidemiology from Spain and Latin America.
Repeated exposome data analysis tutorial
This resource is composed of a Github repository where we provide R codes and simulated data to compare the performance of different statistical approaches to detect exposome-health associations in the context of multiple exposure variables repeatedly assessed in time (longitudinal data).
Implementation of outcome-wide analysis methods tutorial
This includes a tutorial, scripts, and a simulated multivariate dataset for running outcome-wide analysis methods in the context of the exposome, and we explain steps including data standardization, model parameters calibration, output curation strategies, and output visualization.
Federated Lasso for feature-selection with the control of covariates in DataSHIELD (dsLassoCov) tutorial
This tutorial presents a showcase with real cohort data from the HELIX project illustrating how to perform feature selection with regularized regression methods (LASSO and ElasticNet) in DataSHIELD.
Privacy protected federated omic data analysis in multi-center studies with DataSHIELD tutorial
This tutorial is on the use of OmicsSHIELD to conduct privacy protected federated omic data analysis in multi-center studies with DataSHIELD.
DataSHIELD beginners’ workshop
The beginners’ workshop covers the basics of how to begin to use DataSHIELD statistical software. The first part focuses on getting the training environment set up, followed by working through several sections on various statistical techniques.
DataSHIELD resources workshop
This workshop covers how to use the resource, a DataSHIELD extension that allows to manage any large data sets.
DataSHIELD OMICs analysis workshop
This workshop covers how to perform state‐of‐the‐art methods in omic data using non‐disclosive approaches implemented in DataSHIELD including GWAS, gene expression and methylation data analysis.
Statistical analysis to study exposome workshop
This workshop provides a brief overview of available statistical methods to study exposome‐health outcome associations. We explain the data pre‐processing steps required to perform such analysis, and introduce the rexposome package (ExWAS, data visualization).
Tutorial: Non-disclosive federated exposome data analysis with DataSHIELD and Bioconductor
This R package allows us, among others, the analysis of exposome data since we can define a resource as an ExposomeSet an specific method to encapsulate exposome data in R.
ShinyDataSHIELD—an R Shiny app to perform federated non-disclosive data analysis in multicohort studies
ShinyDataSHIELD is a web application with an R backend that serves as a graphical user interface (GUI) to the DataSHIELD infrastructure.
exposomeShiny—a toolbox for exposome data analysis
ExposomeShiny is a standalone web application implemented in R. It is available as source files and can be installed in any server or computer avoiding problems with data confidentiality. It is executed in RStudio which opens a browser window with the web application.
Longitudinal exposome evidence
Web application to query and download findings from Exposome-omics-Wide Association Study (ExWAS)
Our comprehensive and unique resource of exposome-omics associations that serves to replicate findings from other studies and to guide future investigation into the biological imprints of the early life exposome.
Catalogue “blood eQTMs in children”
Web application to download correlations between DNA methylation and expression of nearby genes (eQTMs) in child blood. It will help biological interpretation of epigenome-wide association studies (EWAS) in children.
Click here to access the web application with downloadable files at the end
Machine learning code to compute health environmental-clinical risk scores in European children
In this notebook, Python files are provided to compute Environmental-Clinical risk scores (ECRS) for three main outcomes: behavioural problems (p-factor), metabolic syndrome and lung function. To reference: Guimbaud JB et al. (2024). Machine learning-based health environmental-clinical risk scores in European children. Communications Medicine 4(98).
Knowledge translation tools
Briefing on urban health solutions
This briefing highlights four interventions, which are considered effective at reducing harmful urban exposures and improving health, according to expert consensus surveys and workshops carried out by the ATHLETE project.
Briefing on tackling endocrine disrupting chemicals
The briefing encourages policymakers to introduce EU, national, and local policies targeting the use of chemicals in processing, manufacturing, and packaging without delay.
Plausibility database
This database identifies evidence regarding the effects of chemical and urban factors on child health and attribute a level of evidence (LoE) for each factor-outcome pair. To reference: “Colzin S et al. (2024). A plausibility database summarizing the level of evidence regarding the hazards induced by the exposome on children health. International Journal of Hygiene and Environmental Health 256:114311.” Most recent version: v1.2 (25 Nov. 2023).
Dose-response relationships in children – European exposure levels
The dose-response relationships that you can find in this file are from the systematic review of Rocabois et al., which focused on chemical exposures occuring during pregnancy and childhood and health outcomes occuring before 18th birthday. To reference: Rocabois A et al. (2024) Chemical exposome and children health: identification of dose-response relationships from meta-analyses and epidemiological studies. Environmental Research 119811.
Webinar: Prenatal exposure to phthalates, bisphenol, and organophosphate pesticide mixtures and fetal growth
In collaboration with ATHLETE partner Health and Environment Alliance (HEAL), Dr. Kelly Ferguson (NIEHS) presents her work on prenatal exposure to nonpersistent chemical mixtures and fetal growth.