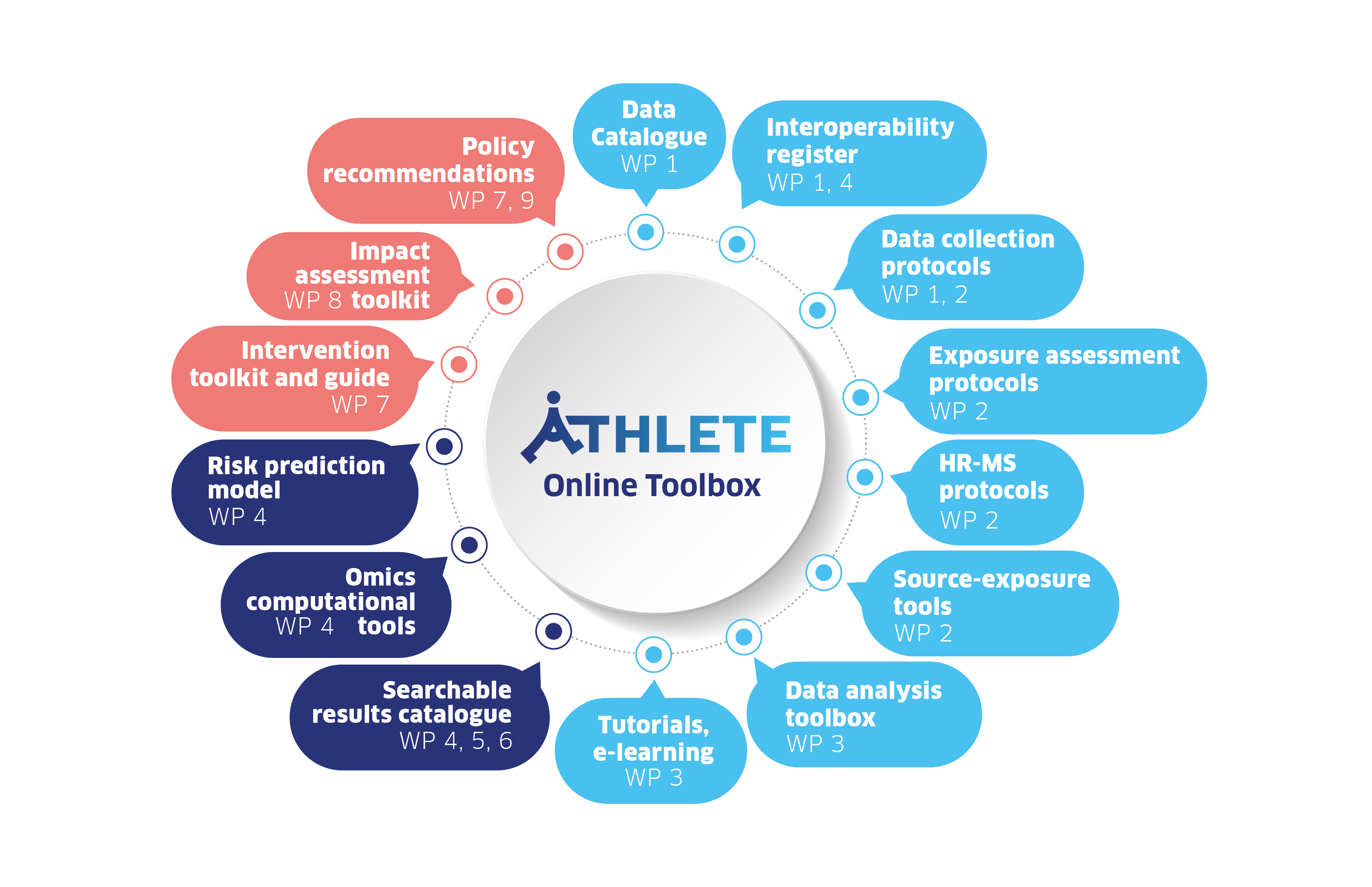
Toolbox
Our online toolbox brings together key data and tools from the ATHLETE project, ensuring they remain available for researchers and policymakers long after the project ends.
By harmonising exposome data from 18 cohorts and making that data easily findable in a catalogue, the ATHLETE project establishes a FAIR (findable, accessible, interoperable, reusable) data infrastructure with a safe and robust data management platform.
HELIX cohort
HELIX cohort
The Human Early Life Exposome (HELIX) cohort study gathers longitudinal population-based birth cohort data from six European countries. It includes biomarkers, omics signatures, and health outcomes. Researchers can get access to this data upon request.
Exposome data
European Health Research Data and Sample Catalogue
This catalogue contains metadata from multiple studies/data sources, their variables, and harmonisation efforts to enable integrated reuse of their rich data. It is a collaborative effort to integrate the catalogues of diverse EU research projects and networks to accelerate data reuse and contribute to improving citizens health.
This video provides details about the catalogue and how it can be used for further research.
Data access protocols
This tool is a technical explanation for the implementation and maintenance of the federated data access system developed by the project. The tool is intended for members of technical teams within institutions or cohorts taking part in or wishing to join the FAIR data infrastructure.
Protocol and SOPs for exposure data, health outcome data, and biological samples collection in adolescents (HELIX cohort)
This tool presents the protocol and standard operating procedures (SOPs) for the collection of data on cardiometabolic, respiratory and neurodevelopment/mental health outcomes, exposures, and biological samples collection for the follow-up of the HELIX cohort into adolescence. This tool is for researchers looking for guidance on cohort data collection protocols aimed at adolescents.
Exposure assessment
Exposure assessment protocols
This document describes the protocols for obtaining personal monitoring data in cohort and intervention study subjects developed during the project, on personal air pollution, physical activity, sleep, and mobility. It also includes protocols for assessing the external exposome using geospatial modelling.
EXPOApp3 and EXPOHub for personal monitoring of physical activity and localisation
The smartphone app ExpoApp3 and its software platform EXPOHub were developed for use within ATHLETE by SME Bettair Cities. The app monitors location (GPS), physical activity, and screen status (on/off) in real-time. It also has an accelerometer. The app can be used alone or as part of a larger set of data collection tools to record personal exposure data in epidemiological and intervention studies.
Analysis tools
Workshop series: Multi-omics data integration in human exposome studies
In this series of virtual workshops, we introduced novel designs and analytical approaches for multi-omics integration that were recently developed and applied in the exposomics field to address critical data analysis challenges, e.g. dimensionality reduction approaches, mediation, predictive modelling, feature selection, networks. The focus is on the practical application of these methods.
Tutorial: Exposome data analysis
A collaborative project combining both plain text explanations and R code chunks for running analyses. Explanations include different exposome analysis steps, namely 1) descriptive analysis, 2) visualization, and 3) association analysis.
Tutorial: Exposome data analysis (in Spanish) – Estrategias para el análisis de datos de Exposoma
A collaborative project combining both plain text explanations and R code chunks for running analyses. Explanations include different exposome analysis steps, namely 1) descriptive analysis, 2) visualization, and 3) association analysis. This material is in Spanish and targeted to researchers in environmental epidemiology.
Tutorial: Repeated exposome data analysis
This resource is composed of a Github repository where we provide R codes and simulated data to compare the performance of different statistical approaches to detect exposome-health associations in the context of multiple exposure variables repeatedly assessed in time (longitudinal data).
Tutorial: Implementation of outcome-wide analysis methods
This resource includes a tutorial, scripts, and a simulated multivariate dataset for running outcome-wide analysis methods in the context of the exposome. The steps including data standardisation, model parameters calibration, output curation strategies, and output visualization are explained.
Tutorial: Federated lasso for feature-selection with the control of covariates in DataSHIELD (dsLassoCov)
This tutorial presents a showcase with real cohort data from the HELIX cohort illustrating how to perform feature selection with regularized regression methods (LASSO and ElasticNet) in DataSHIELD.
Tutorial: Privacy protected federated omic data analysis in multi-center studies with DataSHIELD
This tutorial covers the use of OmicsSHIELD to conduct privacy protected federated omic data analysis in multi-center studies with DataSHIELD. This material is intended to be a quick reference guide for new researchers interested in this technology. It also serves as an online companion for the publication – Federated privacy-protected meta- and mega-omics data analysis in multi-center studies with a fully open-source analytic platform.
Workshop: DataSHIELD for beginners
This beginners workshop covers the basics of how to begin to use the DataSHIELD statistical software. The first part focuses on getting the training environment set up, followed by working through several sections on various statistical techniques. Target audience: Newcomers and beginners to DataSHIELD statistical software.
Workshop: DataSHIELD resources
This workshop covers how to use resources in DataSHIELD, an extension that allows for management of large data sets. Target audience: Researchers interested in more advanced features of DataSHIELD.
Workshop: DataSHIELD OMICs analysis
This workshop covers how to perform state‐of‐the‐art methods with omics data using non‐disclosive approaches implemented in DataSHIELD including GWAS, gene expression and methylation data analysis.
Workshop: Statistical analysis to study the exposome
This workshop provides a brief overview of available statistical methods to study exposome‐health outcome associations. The R exposome package (ExWAS and data visualisation) is explained, including the data pre‐processing steps required to perform such an analysis.
Tutorial: Non-disclosive federated exposome data analysis with DataSHIELD and Bioconductor
This R package allows for the analysis of exposome data in DataSHIELD, by facilitating the use of the resource ExposomeSet, as a specific method to encapsulate exposome data in R.
ShinyDataSHIELD – an R Shiny app to perform federated non-disclosive data analysis in multicohort studies
ShinyDataSHIELD is a web application with an R backend that serves as a graphical user interface (GUI) to the DataSHIELD infrastructure.
exposomeShiny – a toolbox for exposome data analysis
ExposomeShiny is a standalone web application implemented in R. It is available as source files and can be installed in any server or computer avoiding problems with data confidentiality. It is executed in RStudio which opens a browser window with the web application and provides a bridge between researchers and most of the state-of-the-art exposome analysis methodologies, without the need for advanced programming skills.
Longitudinal exposome evidence
Web application to query and download findings from exposome-outcome-wide association study
This web application can be used to query and download findings from the Outcome-Wide Exposome Association Study conducted in the Human Early Life Exposome (HELIX) project, including 586 mother-child pairs from 6 European cohorts in the UK, France, Spain, Lithuania, Norway and Greece, with data on environmental exposures during pregnancy and childhood (6-11 years), and health outcomes covering 4 health domains (mental, cardiometabolic, respiratory, allergy) during childhood (6-11 years) and adolescence (12-18 years).
Web application to query and download findings from exposome-omics-wide association study (ExWAS)
A comprehensive and unique resource of exposome-omics associations that serves to replicate findings from other studies and to guide future investigation into the biological imprints of the early life exposome.
Catalogue – Blood eQTMs in children
Web application to download correlations between DNA methylation and expression of nearby genes (eQTMs) in child blood. It will help biological interpretation of epigenome-wide association studies (EWAS) in children.
Machine learning code to compute health environmental-clinical risk scores in European children
In this notebook, Python files are provided to compute Environmental-Clinical risk scores (ECRS) for three main outcomes: behavioural problems (p-factor), metabolic syndrome and lung function. Reference: Guimbaud JB et al. (2024). Machine learning-based health environmental-clinical risk scores in European children. Communications Medicine 4(98).
Knowledge translation tools
Reducing urban and chemical exposures: a toolkit for action
This toolkit provides a range of resources for the general public, schools, and policymakers seeking to reduce environmental exposures. It includes guidance on interventions that can be implemented at individual, community, and policy levels, along with recommendations and insights into best practices for implementation to inform decision-making and promote healthier environments.
Plausibility database
This plausibility database summarises the level of evidence regarding the effects of environmental hazards (chemical and urban) linking the exposome's effect on children's health. This assessment of evidence levels can synthesise findings across many exposure factors and outcomes, identify understudied domains in children's environmental health, highlight emerging concerns, or help perform health impact assessment studies.
More information about the approach that was developed to extract and summarise the existing evidence about effects of environmental factors on health, is available in a publication by Colzin S et al. (2024).The plausibility database is available in a downloadable table.
Table of dose-response relationships in children – European exposure levels
The dose-response relationships that you can find in this downloadable file can be found in the systematic review of Rocabois et al.(2024) , which focused on chemical exposures occurring during pregnancy and childhood, and different health outcomes– metabolic, neurodevelopmental, respiratory – occurring before a child‘s 18th birthday. This study is one of the largest systematic reviews on dose-response relationships of chemical exposures focusing on children, in terms of number of exposome components considered and could help perform health impact assessments.